

Stephanie Aktipis Department of Organismic and Evolutionary Biology, Museum of Comparative Zoology, Harvard University
Introduction
Gastropods are one of the most evolutionarily successful groups of animals on the planet, inhabiting all continents except Antarctica and occupying a wide variety of marine, freshwater and terrestrial habitats. Although the evolutionary diversity of gastropods makes them an extremely interesting group to study, researchers have had difficulty obtaining a robust phylogenic hypothesis for this class. My Ph.D. research contributes to the understanding of deep evolutionary splits and major evolutionary patterns among the molluscan class Gastropoda, especially those once placed in Archaeogastropoda: Patellogastropoda, Vetigastropoda, Neritimorpha, and Neomphalina. Early speciation events and high amounts of character homoplasy have made obtaining a robust phylogenetic hypothesis for gastropod evoution difficult; relationships among major gastropod clades vary significantly among analyses based on morphological and molecular data sets. Furthermore, results of molecular analyses deviate as different genes are utilized in molecular analyses (see Aktipis et al. 2008 for a historical overview). One of the most problematic differences in these phylogenetic analyses is that Gastropoda is often not recovered as monophyletic when using molecular data. As a result, a major goal of my research is to further test the phylogenetic relationships among main gastropod clades such as Patellogastropoda, Vetigastropoda, Neritimorpha, Neomphalina, and Apogastropoda by increasing molecular data collection and sampling amongst gastropod taxa.
Research Methodology
As my Ph.D. research primarily involves the analysis of molecular data across many gastropod species, the first component of my research necessarily involved the location and preservation of all the species included in my analyses. I was fortunate to obtain many of these important specimens from colleagues both in the United States and abroad, however I also conducted several field expeditions to collect other specimens. After obtaining the specimens, I first extracted DNA from tissue frozen at -80°C or preserved in EtOH or RNAlater. This DNA was then used to sequence the following genes to generate a total of up to 6kb of molecular data for each specimen: 18S rRNA; 28S rRNA; 16S rRNA; cytochrome c oxidase subunit I (hereinafter COI); and histone H3 (hereinafter H3).
In addition to the data from these genes commonly used in molecular analyses, I generated data from other molecular sources. Specifically, I added additional ‘novel’ nuclear protein coding genes (fragments of the head portion of myosin heavy chain type II and elongation factor-1a) to this ‘standard’ 5-gene data set in order to resolve deep splits among the ‘archaeogastropods.’ The techniques required for sequencing these ‘novel’ genes necessitate the use of specific protocols involving RNA instead of DNA, thereby restricting their application to the limited number of specimens in my collection frozen or preserved in RNAlater.
For my molecular analyses, I generated sequence data through a process of direct sequencing utilizing polymerase chain reaction products on an ABI 3730 automated sequencer. My phylogenetic analysis utilizes multiple methods including parsimony and maximum likelihood approaches. Due to the difficulty of collecting fresh specimens for the RNA extraction and sequencing of the ‘novel’ genes, two independent studies were performed. In one, all seven molecular loci were sequenced from 31 ingroup taxa representing all ‘archaeogastropod’ clades except Cocculinoidea and 8 ougroup taxa. The second study increases in the number of ‘archaeogastropod’ specimens sampled (104 taxa) and included species from all ‘archaeogastropod’ groups but only five genes were sequenced from these specimens.
Results
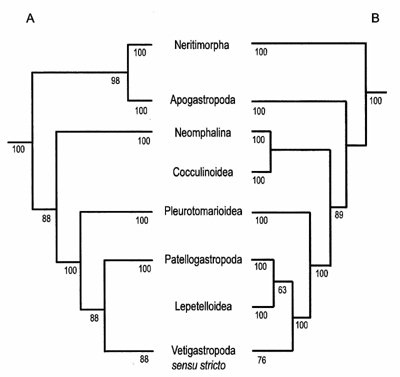 |
Figure 1: Schematic representation of gastropod relationships based on the results of the maximum likelihood trees from the two studies. (A) Summary tree of the maximum likelihood analysis for the 7-gene combined molecular analysis (-log L = 95,447.3787). The topology of the optimal tree from the parsimony analysis was very similar except Vetigastropoda was not monophyletic and Neomphalina was sister to Pleurotomarioidea. (B) Summary tree of the maximum likelihood analysis for the 5-gene combined molecular analysis (-log L = 170,159.1065). The topology of the optimal tree from the parsimony analysis was identical except for the internal relationships among groups within Vetigastropoda sensu stricto. Numbers on branches indicate bootstrap support values above 50%. |
The second study, “Relationships among the vetigastropod taxa: a molecular approach” investigates the internal relationships among putative members of the clade Vetigastropoda (Fissurelloidea, Haliotoidea, Lepetelloidea, Lepetodriloidea, Pleurotomarioidea, Scissurelloidea, Seguenzioidea, Trochoidea, Turbinoidea, Neomphaloidea, and Cocculinoidea) in a molecular phylogeny utilizing nearly six kilobases of molecular data from up to five nuclear and mitochondrial genes. This study includes sequence data from all vetigastropod clades and represents an almost fourfold increase in the number of vetigastropod specimens as compared to the more limited vetigastropod taxon, yet higher sampling of genetic loci, in the first study. In order to overcome shortfalls of previous vetigastropod analyses resulting from the under-sampling of outgroups, this study includes broad outgroup representation. Fissurelloidea, Haliotoidea, Lepetodriloidea, Scissurellidae, Seguenzioidea, Trochoidea, and Turbinoidea formed a clade identified as Vetigastropoda sensu stricto united by morphological synapomorphies such as the presence of bursicles and epipodial sense organs. In contrast, Neomphalina, Cocculinoidea, and Pleurotomarioidea fell outside of Vetigastropoda sensu stricto, indicating a need to recognize these clades as independent of vetigastropods (Figure 1B). The placement of Neophalina, Cocculinoide and Pleurotomarioidea outside of Vetigastropoda sensu strico was consistent in both the parsimony and likelihood analyses, but the relationships among the groups within Vetigastropoda sensu stricto varied among the analyses performed.
The phylogenetic placement of major ‘archaeogastropod’ clades in both studies was remarkably similar. In the optimal parsimony and maximum likelihood analyses for both studies, Neritimorpha fell outside all remaining ‘archaeogastropods’ and Vetigastropoda sensu stricto was sister to Patellogastropoda. Furthermore, Neomphalina, Cocculinoidea, and Pleurotomarioidea were independent groups outside the Vetigastropoda + Patellogastropoda clade. Comparing the results of these studies indicates that increasing characters as well as taxa in phylogenetic analyses improves both the resolution and stability of phylogenetic hypotheses.
I thank the Malacological Society of London for their generous financial support that made a significant portion of this phylogenetic project possible.
References
Aktipis, S. W., Giribet, G., Lindberg, D. R., and Ponder, W. F. (2008). Gastropod Phylogeny: an overview and analysis. In Phylogeny and Evolution of the Mollusca. D. R. Lindberg and W. F. Ponder. Berkeley, CA: Berkeley University Press: 201-237.